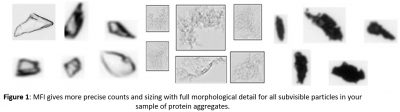
Sujet : « Biopharmaceutical particle classification and characterization through deep-learning inside a syringe »
Keywords
machine learning, deep tech, VAE, characterization method, micro-Flow Imaging, coating layer thickness, thin coating characterization
Context and objectives
Micro-flow imaging (MFI) is a sensitive, simple and automated method for the analysis of sub-visible particles and translucent protein aggregates that provides particle size, count and morphology. Pharmacopeial agencies are demanding more information on the nature of particles within a pharmaceutical product.
Mechanical, chemical and topographic characterization of regular or crosslinked silicone oil (PDMS) inside a glass barrel – the syringe – is very challenging [4]. Particles are potentially present in all biopharmaceutical samples and can originate from various sources: production process, formulation excipients, degradation products, particle impurities, etc. Particles occur in various sizes.
The presence of particles can on one hand indicate an insufficient stability of the active pharmaceutical ingredient (API) or issues with the production process, on the other hand, can negatively influence the safety and efficacy of the therapy. This is why is particle characterization is so crucial.
Most characterization methods are time-consuming, have limited access inside the barrel, are destructive or not scalable [1]. For example, some current practices (e.g. standard AFM) limit characterization to few samples in a week duration for preparation, calibration and measurement.
Modeling by statistical learning is one of the possible solutions, with the undeniable advantage of being able to model massive amounts of data and to discover high-level representations [2]. Deep learning quickly established itself as a standard in several fields, smashing the records for various state-of-the-art methods [2]. In this project we will focus more specifically on the classification of particles with the objective of processing more than 35,000 images per minutes. These methods also require numerous annotated examples to form a model. This project addresses the problem of transfer learning and data augmentation in the context of insufficient data.
Methods
Deep learning analysis of MFI (Micro-Flow Imaging) data is almost non-existent. The reason is the lack of large MFI particles data sets available due to their cost of acquisition and variability in size, composition, etc. The difficulty of obtaining a sufficient amount of reliable class-specific training data for a supervised automatic approach requires the study of new strategies. A solution suggested by very recent studies [3], proposes to develop new generic salience functions or to use the data augmentation method to build a robust classification as well as other parameters such as texture or shape. Learning by neural networks inspired by ladder networks or regime networks or adversary autoencoding, curicular model, etc. will be privileged in this internship.
The solution to the issue is to segment particles suspended in the fluid by machine learning (to overcome traditional image processing segmentation techniques) and for this (a) the algorithm would be based largely on the already available MFI imaging data mentioned (b) will also take into account multi-modal data integrating the particle size distribution, etc.
There are 2 levels of analysis for this problem: automatic classification of particles or quantification of the fluid as a whole. Both are accessible.
The proposed classification algorithm should be more discriminating than those used today on MFI images, while allowing rapid analysis of the particles present on the images, the flow rate of MFI machines being very high today, around 200 µl / min with a concentration of 175,000 particles / ml).
Thus, based on the information shared at this stage, two algorithmic approaches seem interesting to us:
– use deep learning on a USR (Ultrafast Shape Recognition) system;
– adapt modern image recognition neural networks dedicated to edge computing known to be light and not very CPU intensive.
Numerous publications in various disciplinary fields (physics, geology, medical imaging, etc.) have already demonstrated the effectiveness of multimodality for machine learning.
Note that the physical characteristics mentioned in the subject (size, diameter, height / width ratio, circularity, area, perimeter, intensity) are not necessarily taken into account explicitly because this information is de facto present in the image.
In the absence of ground truth, particle size characterization is essential to verify the performance of the program and the quality of the segmentation.
Program of work
step 1: Collect existing data step
step 2: Feasibility. Prototype of a neural network model. 2. Network training on the base created in “step 1”.
Expected results
The creation of a classifier capable of predicting and discriminating similar shapes in low resolution images for objects of size [5µ, 70 µ]. Classes could be glass, cellulose, protein mimics and lysozyme aggregate.
The expected performance indicators are: (a) the precision of the predictions (b) the repeatability of the prediction process (c) the robustness in degraded situations (d) the satisfaction of the quality officer
Potential decision/performance criteria are: identification time, classification performance indices (sensitivity, false positives, etc.), reproducibility of the results
Profile and skills required
Ability to understand and develop adaptive learning algorithms and to process medical data, index it and use it in an operational system to achieve the mission described above.
Programming skills: Python or C / C ++. A practice of Tensorflow and Pytorch would be a plus. The practice of French is not compulsory.
Professional qualities sought
autonomy, sense of relationship to interact with research and company teams, motivation for new technologies, creativity to set up an innovative solution.
Supervision and scientific and material conditions
The project is multidisciplinary, at the interface of machine learning, computer science and physics. The student will be supervised by Vincent Vigneron, Jean-Philippe Congé and Hichem Maaref from the IBISC laboratory (Univ d’Évry, Université Paris-Saclay). All master machine learning, signal and image processing.
Contact
Vincent Vigneron & Hichem Maaref {hichem.maaref,vincent.vigneron}@univ-evry.fr
Phone: +33 6 635 687 60
Start and end date of the internship: 15/02/2022 au 1/09/2022
References
- [1] Nayyer Aafaq, Ajmal Mian, Wei Liu, Syed Zulqarnain Gilani, and Mubarak Shah. 2019. Video description: A survey of methods, datasets, and evaluation metrics. ACM Computing Surveys (CSUR) 52, 6 (2019), 1–37.
- [2] LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning, Nature, 521(7553) :436–444.
- [3] Obeso, A. M., Benois-Pineau, J., Guissous, K., Gouet-Brunet, V., García Vázquez, M. S., and Ramírez Acosta, A. A. (2018). Comparative study of visual saliency maps in the problem of classification of architectural images with deep CNNs. In 2018 Eighth International Conference on Image Processing Theory, Tools and Applications (IPTA), pages 1–6.
- [4] J. Houde, A.S. Berkowitz, eds., Biophysical Characterization of Proteins in Developing Biopharmaceuticals, 1st ed., Newnes, 2014
- Date de l’appel : 13/12/2021
- Statut de l’appel : Pourvu
- Contacts cotés IBISC : Jean-Philippe CONGE (doctorant IBISC, équipe SIAM), Vincent VIGNERON (MCF HDR Univ. Évry, IBISC équipe SIAM), Hichem MAAREF (PR IUT Évry, IBISC équipe SIAM) {vincent.vigneron,hichem.maaref}@ibisc.univ-evry.fr, }
- Sujet de stage niveau Master 2 (format PDF)
- Web équipe SIAM