Intership title : AI characterization of the MR SWI-DWI ischemic mismatch for hyperacute ischemic stroke patients
Partners: IBISC (Université d’Évry, Université Paris-Saclay), Centre Hospitalier Sud
Francilien (CHSF), Centre Hospitalier Régional Universitaire de Tours (CHRU)
Basic AI and Data Science: statistical training in big dimensions
Specialized ML and AI: signal, image, vision
Application domain: precision medicine, imagery by MR
Mots-clés deep learning, multi-modality imaging, semi-supervised learning
Key-words machine learning, deep tech, neuroimaging, precision medicine, stroke
Total duration of internship: 6 months (postgraduate) Working period: From 2025/02/01 to 2025/09/01
Context and objectives
According to the World Health Organization, stroke is the second cause of death, the first for women, and the leading cause of chronic functional disability in adults, with 17 million victims, 31% of whom were under 65.
In France, around 150,000 people are hospitalized yearly for a stroke, one every 4 minutes. It represents a financial burden of about 2.8 billion €/year; in reality, 10 billion over five years due to the cost of disability.
Ischemic stroke is caused by a blood clot (thrombus) that blocks a brain artery, causing a lack of oxygen to brain tissue supplied by that artery. There is an urgent need to diagnose and determine the choice of treatment.
Neuroimaging plays a decisive role in demonstrating acute ischemia with the concept of ischemic penumbra. It has been shown that the growth, duration, and infarct volume of ischemic stroke directly influence the amount of irreversible tissue loss. There is a need for a quantitative penumbra-assessment stroke protocol with the minimum possible imaging time that delivers the maximum possible information. Today, this assessment is made with perfusion imaging, with the risk of exposure to radiation and use ofiodinated contrast medium, which may affect renal-impaired patients. For this reason, finding other solutions have been a focus of research. An MR-based fast, non-invasive stroke protocol such as T2* gradient-echo and susceptibility-weighted imaging (SWI) are proposed solutions in the literature to measure the penumbra quantitatively.
Objectives
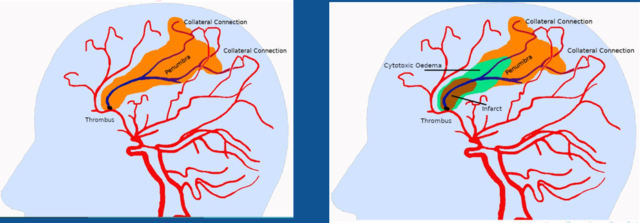
The solution we want to implement is based on automatically segmenting the area of already dead tissue (infarct) and ischemic tissues at risk (penumbra), as seen in Figure 1. The penumbra assessment can be obtained through the mismatch ratio obtained from the MR perfusion-weighted imaging (PWI) and the diffusion-weighted imaging (DWI) modalities. Recent studies have proposed a framework for accurate quantitative assessment of penumbra using SWI-DWI and its validation with PWI-DWI-based quantification [1].
These assessments depend heavily on the user expertise. The current study aims to develop an automated image-processing framework for quantifying penumbral volume using SWI and/or DWI [2]. Applying AI algorithms to the analysis of MR images makes it possible to work on large amounts of data in a more relevant way than conventional statistical methods. The objectives are (1) to validate the results on an extensive patient database (2) to integrate the model into clinical application software with a user-friendly interface. The current study also includes the study of the penumbra volume as a prognosis tool in the case of revascularization.
Methodology
No automatic method of quantification of penumbra volume using SWI and DWI has been published to date. A few publications are on semi-automatic segmentation of the lesion by angiography [4, 10, 7, 12, 11]. Only the last three relate to the segmentation of the lesion in the brain. These studies all have in common that they need a “manual” seed to run the algorithm, meaning they cannot find the position of the lesion on their own.
Expected results
The expected solution will better characterize the ischemic mismatch by associating multiple weak MRI signals with the definition of pathology. The MR mismatch will improve the reading quality of current images by performing analyses that are not currently carried out because they take too long to execute manually, such as the volumetric measurement of the penumbra. Moreover, the MR mismatch solution without a contrast agent will make this analysis quicker and more available to all types of patients.
This solution will determine what information in the image implies specific treatments leading to a better patient prognosis. AI can help the radiologist prioritize urgent cases by deciding which imaging tests to assess first. Ultimately, radiology experts should give more information than the human eye on the texture of the thrombus and its accessibility for recanalization treatments.
Expected performance criteria:
Evaluating the new procedure against a referenced approach raises many methodological difficulties. The expected performance indicators are (1) the repeatability of the deterministic segmentation process in a degraded situation or not, (2) the efficiency of the tool to be tested on a ground truth basis and quantified with DICE [3] to measure performance in segmentation, (3) a speed of execution of a few minutes.
References
[1] Bhattacharjee R, Gupta RK, Das B, Dixit VK, Gupta P, Singh A. Penumbra quantification from MR SWI-DWI mismatch and its comparison with MR ASL PWI-DWI mismatch in patients with acute ischemic stroke. NMR Biomed. 2021 Jul;34(7):e4526. doi: 10.1002/nbm.4526. Epub 2021 Apr 20. PMID: 33880799.
[2] Yamaguchi S, Horie N, Morikawa M, Tateishi Y, Hiu T, Morofuji Y, Izumo T, Hayashi K, Matsuo T. Assessment of veins in T2*-weighted MR angiography predicts infarct growth in hyperacute ischemic stroke. PLoS One. 2018 Apr 4;13(4):e0195554. doi: 10.1371/journal.pone.0195554. PMID: 29617449; PMCID: PMC5884555.
[3] Brahim, D. Fourer, V. Vigneron and H. Maaref, « Deep Learning Methods for MRI Brain Tumor Segmentation: a comparative study, » 2019 Ninth International Conference on Image Processing Theory, Tools and Applications (IPTA), 2019, pp. 1-6, doi: 10.1109/IPTA.2019.8936077.
[4] Egger, J., O’Donnell, T., Hopfgartner, C., Freisleben, B. (2009). Graph-based Tracking Method for Aortic Thrombus Segmentation. In: Vander Sloten, J., Verdonck, P., Nyssen, M., Haueisen, J. (eds) 4th European Conference of the International Federation for Medical and Biological Engineering. IFMBE Proceedings, vol 22. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-89208-3_139
[5] Kobold, J. Vigneron, V. Maaref, H. Fourer, D., Aghasaryan, M. Alecu, C. Chausson, N. L’hermitte, Y. Smadja, D. and Läng, E. Stroke Thrombus Segmentation on SWAN with MultiDirectional U-Nets. In 9th IEEE International Conference on Image Processing Theory, Tools and Applications (IPTA 2019), Istanbul, Turkey, November 2019.
[6] Maier O, Menze BH, von der Gablentz J, Ḧani L, Heinrich MP, Liebrand M, Winzeck S, Basit A, Bentley P, Chen L, Christiaens D, Dutil F, Egger K, Feng C, Glocker B, Götz M, Haeck T, Halme HL, Havaei M, Iftekharuddin KM, Jodoin PM, Kamnitsas K, Kellner E, Korvenoja A, Larochelle H, Ledig C, Lee JH, Maes F, Mahmood Q, Maier-Hein KH, McKinley R, Muschelli J, Pal C, Pei L, Rangarajan JR, Reza SMS, Robben D, Rueckert D, Salli E, Suetens P, Wang CW, Wilms M, Kirschke JS, Kr Amer UM, Münte TF, Schramm P, Wiest R, Handels H, Reyes M. ISLES 2015 – A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med Image Anal. 2017 Jan;35:250-269. doi: 10.1016/j.media.2016.07.009. Epub 2016 Jul 21. PMID: 27475911; PMCID: PMC5099118.
[7] Olabarriaga SD, Rouet JM, Fradkin M, Breeuwer M, Niessen WJ. Segmentation of thrombus in abdominal aortic aneurysms from CTA with nonparametric statistical grey level appearance modeling. IEEE Trans Med Imaging. 2005 Apr;24(4):477-85. doi: 10.1109/tmi.2004.843260. PMID: 15822806.
[8] Parmar, C., Grossmann, P., Bussink, J. et al. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 5, 13087 (2015). https://doi.org/10.1038/srep13087
[9] Pustina D, Coslett HB, Turkeltaub PE, Tustison N, Schwartz MF, Avants B. Automated segmentation of chronic stroke lesions using LINDA: Lesion identification with neighborhood data analysis. Hum Brain Mapp. 2016 Apr;37(4):1405-21. doi: 10.1002/hbm.23110. Epub 2016 Jan 12. PMID: 26756101; PMCID: PMC4783237.
[10] Qazi S, Qazi E, Wilson AT, McDougall C, Al-Ajlan F, Evans J, Gensicke H, Hill MD, Lee T, Goyal M, Demchuk AM, Menon BK, Forkert ND. Identifying Thrombus on Non-Contrast CT in Patients with Acute Ischemic Stroke. Diagnostics (Basel). 2021 Oct 16;11(10):1919. doi: 10.3390/diagnostics11101919. PMID: 34679617; PMCID: PMC8534393.
[11] Riedel CH, Jensen U, Rohr A, Tietke M, Alfke K, Ulmer S, Jansen O. Assessment of thrombus in acute middle cerebral artery occlusion using thin-slice nonenhanced Computed Tomography reconstructions. Stroke. 2010 Aug;41(8):1659-64. doi: 10.1161/STROKEAHA.110.580662. Epub 2010 Jul 1. PMID: 20595670.
[12] Santos EM, Marquering HA, Berkhemer OA, van Zwam WH, van der Lugt A, Majoie CB, Niessen WJ; MR CLEAN investigators. Development and validation of intracranial thrombus segmentation on CT angiography in patients with acute ischemic stroke. PLoS One. 2014 Jul 17;9(7):e101985. doi: 10.1371/journal.pone.0101985. PMID: 25032691; PMCID: PMC4102487.
[13] Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuroimage Clin. 2017 Jun 13;15:633-643. doi: 10.1016/j.nicl.2017.06.016. PMID: 28664034; PMCID: PMC5480013.
[14] Dice, L.R. Measures of the amount of ecologic association between species. Ecology, 26(3):297–302, 1945.
Profile and skills required
Ability to understand and develop adaptive learning algorithms and process medical data, index it, and use it in an operational system to achieve the abovementioned mission. Programming skills: Python or C / C ++. A practice of Tensorflow and Pytorch would be a plus. The practice of French is not compulsory. His(her) English is fluent. The work will be done at the IBISC Laboratory on the Evry campus of the Université Paris-Saclay. IBISC develops multidisciplinary, theoretical, and applied research in information sciences and engineering, with a strong orientation towards health applications. The selected candidate will be integrated into an interdisciplinary team with a consortium of data scientists and clinicians from the CHSF and the CHRU in Tours. The project is multidisciplinary, at the interface of machine learning, computer science, and medicine.
Scientific and material conditions
The student will be supervised by Mariana Brejo, Hichem Maaref, and Vincent Vigneron from the IBISC laboratory (Université Évry Paris-Saclay). All master machine learning, signal, and image processing.
Contact: Vincent Vigneron, Hichem Maaref and Mariana Brejo{mariana.goncalvesbrejo,hichem.maaref,vincent.vigneron}@univ-evry.fr, Phone: +33 1 694 775 45
- Date de l’appel : 19/11/2024
- Statut de l’appel : Pourvu
- Contacts cotés IBISC : Vincent VIGNERON (PR Univ. Évry, IBISC équipe SIAM), Hichem MAAREF (PR IUT Évry, IBISC équipe SIAM} et Mariana BREJO (doctorante IBISC équipe SIAM){mariana.goncalvesbrejo,hichem.maaref,vincent.vigneron}@univ-evry.fr
- Sujet de stage niveau Master 2 (format PDF)
- Web équipe SIAM