Thesis topic: Recanalisation prediction for stroke patients
Context
According to the World Health Organization, stroke is the second cause of death, the first for women, and the leading cause of chronic functional disability in adults, with 17 million victims. In France, around 150,000 people are hospitalized yearly for a stroke, one every 4 minutes. It represents a financial burden of about 2.8 billion €/year; in reality, 10 billion over five years due to the cost of disability. 1 over 6 people will have a stroke in their lives.
Ischemic stroke is caused by a blood clot (thrombus) that blocks a brain artery, causing a lack of oxygen to brain tissue supplied by that artery. There is an urgent need to diagnose and determine the choice of treatment. It produces damage in the brain (lesion) in which some parts of the tissue can be recovered (penumbra) and another is dead (core). Localizing the cause (thrombus) and the consequence (lesion) is key to choosing the treatment. Its recovery depends on several factors: delay, type of treatment, diseases, age, size of lesion and clot, etc. Neurologists make the decision thanks to magnetic resonance imaging (MRI). The clot and the lesion are located in several modalities where different information is available. Deep learning techniques could optimize this long procedure.
Objectives
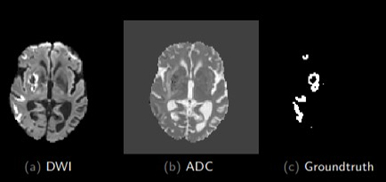
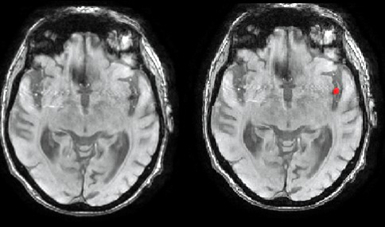
The final objective is to do a deep learning model that helps to make the treatment decision. To do that, the recanalization prediction should be predicted. If a patient is going to recanalize thanks to the usual treatment (thrombolysis), no other treatment is needed, and its use is recommended. To obtain that, useful information from the MRI should be extracted. In Figure 1 we can see the segmentation of the lesion and in Figure 2 the one of the thrombus. An automatic segmentation model based on deep learning is needed to obtain this segmentation with the highest Dice possible without missing any patients. Once this information is extracted, the treatment outcome could be predicted.
Methodology and expected results
There are several possible approaches: once Transformers segment the lesion and thrombus [1] or CNN-based architecture [2], some features from the image could be extracted. These features can be inputs for easy models: logistic regression/random forest [3]. Another possible solution is to directly predict the outcome from the image: how the lesion and the thrombus evolve, for example, [4]. The Dice score is used to compare the models. For that, some public and private datasets will be used in multimodal 3D images with segmentation task.
Profile and skills required
Ability to understand and develop adaptive learning algorithms and process medical data, index it, and use it in an operational system to achieve the above-mentioned mission. Programming skills: Python and Linux language. A practice of Tensorflow and Pytorch would be a plus. Basic knowledge of computer vision is needed, and some experience with a real problem would be a plus.
The practice of French is not compulsory. The work will be done at the IBISC Laboratory on the Evry campus of the Universit´e Paris-Saclay. IBISC develops multidisciplinary, theoretical, and applied research in information sciences and engineering, with a strong orientation towards health applications. The selected candidate will be integrated into an interdisciplinary team with a consortium of data scientists and clinicians from the CHSF. The project is multidisciplinary, at the interface of machine learning, science, and medicine. Several datasets as computer servers are available for the project.
Scientific and material conditions
Sofia Vargas Ibarra and Vincent Vigneron will supervise the students from the IBISC laboratory (Universit´e Paris-Saclay, campus d’Evry). All master machine learning, signal, and image processing.
Contact
Send resume and grades of bachelor+master to Vincent Vigneron and Sofia Vargas Ibarra. {sofia.vargasibarra,vincent.vigneron}@univ-evry.fr
References
[1] Alexey Dosovitskiy, Lucas Beyer, Alexander Kolesnikov, Dirk Weissenborn, Xiaohua Zhai, Thomas Unterthiner, Mostafa Dehghani, Matthias Minderer, Georg Heigold, Sylvain Gelly, Jakob Uszkoreit, and Neil Houlsby. An image is worth 16×16 words: Transformers for image recognition at scale, 2021.
[2] Sofia Vargas Ibarra, Vincent Martin VIGNERON, Sonia Garcia Salicetti, Hichem Maaref, Jonathan Kobold, Nicolas Chausson, Yann Lhermitte, and Didier Smadja. A recurrent network for segmenting the thrombus on brain MRI in patients with hyper-acute ischemic stroke. In Medical Imaging with Deep Learning, 2024.
[3] Young Dae Kim, Hyo Suk Nam, Joonsang Yoo, Hyungjong Park, Sung-Il Sohn, Jeong-Ho Hong, Byung Moon Kim, Dong Joon Kim, Oh Young Bang, Woo-Keun Seo, et al. Prediction of early recanalization after intravenous thrombolysis in patients with large-vessel occlusion. Journal of Stroke, 23(2):244, 2021.
[4] Sanaz Nazari-Farsani, Yannan Yu, Rui Duarte Armindo, Maarten Lansberg, David S Liebeskind, Gregory Albers, Soren Christensen, Craig S Levin, and Greg Zaharchuk. Predicting final ischemic stroke lesions from initial diffusion-weighted images using a deep neural network. NeuroImage: Clinical, 37:103278, 2023.
- Date de l’appel : 10/10/2024
- Statut de l’appel : Pourvu
- Contacts cotés IBISC : Vincent VIGNERON (PR Univ. Évry, IBISC équipe SIAM), Sofia VARGAS IBARRA (doctorante IBISC, équipe SIAM){sofia.vargasibarra,vincent.vigneron}@univ-evry.fr
- Sujet de stage niveau Master 2 (format PDF)
- Web équipe SIAM